Chapter 5 and tidymodels
Cornell College
STA 362 Spring 2024 Block 8
Setup
Cross validation
💡 Big idea
- We have determined that it is sensible to use a test set to calculate metrics like prediction error
Cross validation
💡 Big idea
- We have determined that it is sensible to use a test set to calculate metrics like prediction error
Why?
Cross validation
💡 Big idea
- We have determined that it is sensible to use a test set to calculate metrics like prediction error
How could we do this?
Cross validation
💡 Big idea
- We have determined that it is sensible to use a test set to calculate metrics like prediction error
- What if we don’t have a separate data set to test our model on?
- 🎉 We can use resampling methods to estimate the test-set prediction error
Training error versus test error
What is the difference? Which is typically larger?
- The training error is calculated by using the same observations used to fit the statistical learning model
- The test error is calculated by using a statistical learning method to predict the response of new observations
- The training error rate typically underestimates the true prediction error rate
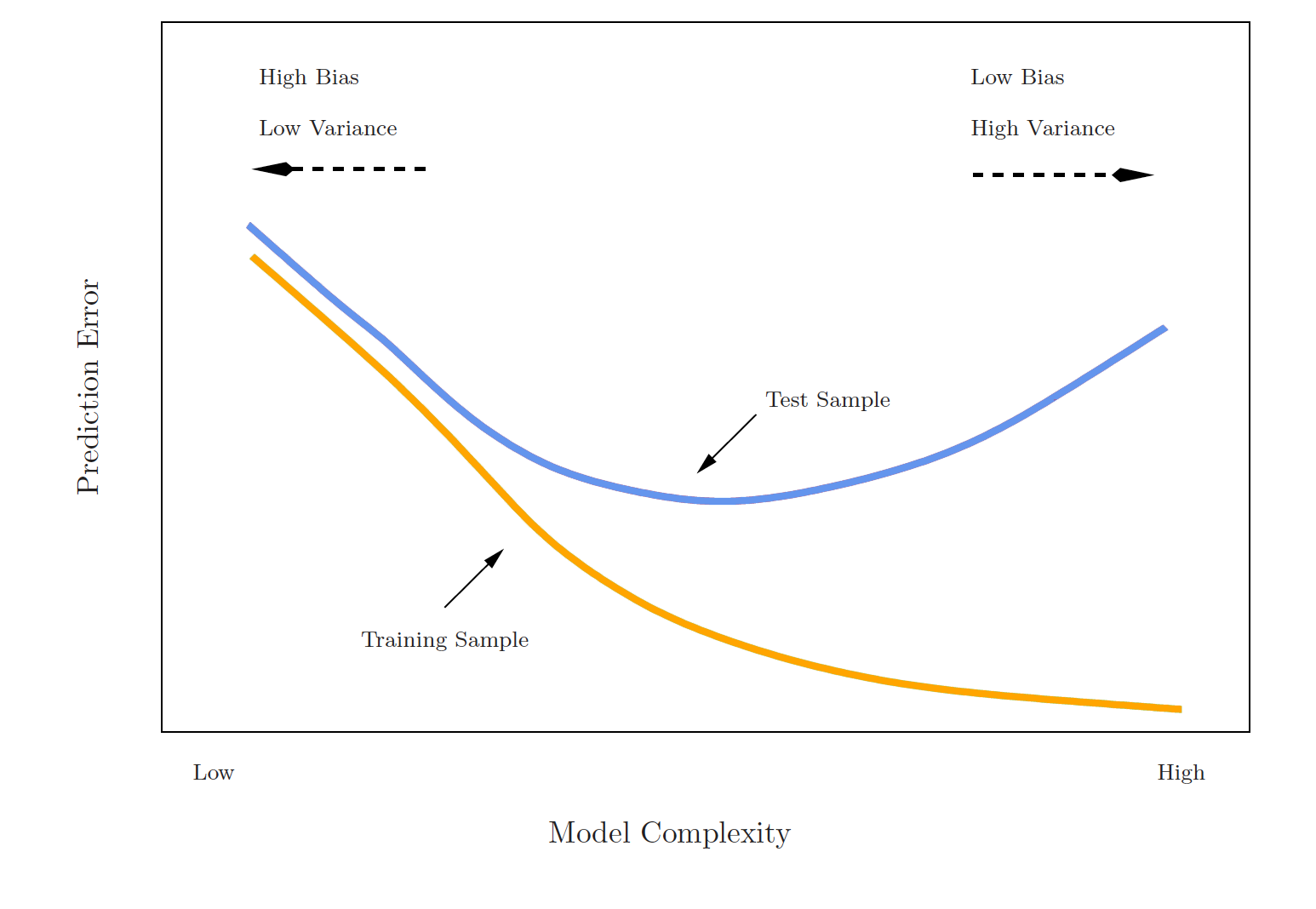
Estimating prediction error
- Best case scenario: We have a large data set to test our model on
- This is not always the case!
💡 Let’s instead find a way to estimate the test error by holding out a subset of the training observations from the model fitting process, and then applying the statistical learning method to those held out observations
Approach #1: Validation set
- Randomly divide the available set up samples into two parts: a training set and a validation set
- Fit the model on the training set, calculate the prediction error on the validation set
If we have a quantitative predictor what metric would we use to calculate this test error?
- Often we use Mean Squared Error (MSE)
Approach #1: Validation set
- Randomly divide the available set up samples into two parts: a training set and a validation set
- Fit the model on the training set, calculate the prediction error on the validation set
If we have a qualitative predictor what metric would we use to calculate this test error?
- Often we use misclassification rate
Approach #1: Validation set

\[\Large\color{orange}{MSE_{\texttt{test-split}} = \textrm{Ave}_{i\in\texttt{test-split}}[y_i-\hat{f}(x_i)]^2}\]
\[\Large\color{orange}{Err_{\texttt{test-split}} = \textrm{Ave}_{i\in\texttt{test-split}}I[y_i\neq \mathcal{\hat{C}}(x_i)]}\]
Approach #1: Validation set
Auto example:
- We have 392 observations.
- Trying to predict
mpgfromhorsepower. - We can split the data in half and use 196 to fit the model and 196 to test
Approach #1: Validation set

\(\color{orange}{MSE_{\texttt{test-split}}}\)

\(\color{orange}{MSE_{\texttt{test-split}}}\)

\(\color{orange}{MSE_{\texttt{test-split}}}\)

\(\color{orange}{MSE_{\texttt{test-split}}}\)
Approach #1: Validation set
Auto example:
- We have 392 observations.
- Trying to predict
mpgfromhorsepower. - We can split the data in half and use 196 to fit the model and 196 to test - what if we did this many times?
Approach #1: Validation set (Drawbacks)
- the validation estimate of the test error can be highly variable, depending on which observations are included in the training set and which observations are included in the validation set
- In the validation approach, only a subset of the observations (those that are included in the training set rather than in the validation set) are used to fit the model
- Therefore, the validation set error may tend to overestimate the test error for the model fit on the entire data set
Approach #2: K-fold cross validation
💡 The idea is to do the following:
- Randomly divide the data into \(K\) equal-sized parts
- Leave out part \(k\), fit the model to the other \(K - 1\) parts (combined)
- Obtain predictions for the left-out \(k\)th part
- Do this for each part \(k = 1, 2,\dots K\), and then combine the result
K-fold cross validation

\(\color{orange}{MSE_{\texttt{test-split-1}}}\)

\(\color{orange}{MSE_{\texttt{test-split-2}}}\)

\(\color{orange}{MSE_{\texttt{test-split-3}}}\)

\(\color{orange}{MSE_{\texttt{test-split-4}}}\)
Take the mean of the \(k\) MSE values
Application Exercise
Create a new R project, then a new quarto file with cv in its name in that project. Answer the questions in that file.
If we use 10 folds:
- What percentage of the training data is used in each analysis for each fold?
- What percentage of the training data is used in the assessment for each fold?
Estimating prediction error (quantitative outcome)
- Split the data into K parts, where \(C_1, C_2, \dots, C_k\) indicate the indices of observations in part \(k\)
- \(CV_{(K)} = \sum_{k=1}^K\frac{n_k}{n}MSE_k\)
- \(MSE_k = \sum_{i \in C_k} (y_i - \hat{y}_i)^2/n_k\)
- \(n_k\) is the number of observations in group \(k\)
- \(\hat{y}_i\) is the fit for observation \(i\) obtained from the data with the part \(k\) removed
- If we set \(K = n\), we’d have \(n-fold\) cross validation which is the same as leave-one-out cross validation (LOOCV)
Estimating prediction error (quantitative outcome)
- Split the data into K parts, where \(C_1, C_2, \dots, C_k\) indicate the indices of observations in part \(k\)
- \(CV_{(K)} = \sum_{k=1}^K\frac{n_k}{n}MSE_k\)
- \(MSE_k = \sum_{i \in C_k} (y_i - \hat{y}_i)^2/n_k\)
- \(n_k\) is the number of observations in group \(k\)
- \(\hat{y}_i\) is the fit for observation \(i\) obtained from the data with the part \(k\) removed
- If we set \(K = n\), we’d have \(n-fold\) cross validation which is the same as leave-one-out cross validation (LOOCV)
Leave-one-out cross validation




\(\dots\)

Special Case!
- With linear regression, you can actually calculate the LOOCV error without having to iterate!
- \(CV_{(n)} = \frac{1}{n}\sum_{i=1}^n\left(\frac{y_i-\hat{y}_i}{1-h_i}\right)^2\)
- \(\hat{y}_i\) is the \(i\)th fitted value from the linear model
- \(h_i\) is the diagonal of the “hat” matrix (remember that! 🎓)
Picking \(K\)
- \(K\) can vary from 2 (splitting the data in half each time) to \(n\) (LOOCV)
- LOOCV is sometimes useful but usually the estimates from each fold are very correlated, so their average can have a high variance
- A better choice tends to be \(K=5\) or \(K=10\)
Bias variance trade-off
- Since each training set is only \((K - 1)/K\) as big as the original training set, the estimates of prediction error will typically be biased upward
- This bias is minimized when \(K = n\) (LOOCV), but this estimate has a high variance
- \(K =5\) or \(K=10\) provides a nice compromise for the bias-variance trade-off
Approach #2: K-fold Cross Validation
Auto example:
- We have 392 observations.
- Trying to predict
mpgfromhorsepower
Estimating prediction error (qualitative outcome)
- The premise is the same as cross valiation for quantitative outcomes
- Split the data into K parts, where \(C_1, C_2, \dots, C_k\) indicate the indices of observations in part \(k\)
- \(CV_K = \sum_{k=1}^K\frac{n_k}{n}Err_k\)
- \(Err_k = \sum_{i\in C_k}I(y_i\neq\hat{y}_i)/n_k\) (misclassification rate)
- \(n_k\) is the number of observations in group \(k\)
- \(\hat{y}_i\) is the fit for observation \(i\) obtained from the data with the part \(k\) removed
Estimating prediction error (qualitative outcome)
- The premise is the same as cross valiation for quantitative outcomes
- Split the data into K parts, where \(C_1, C_2, \dots, C_k\) indicate the indices of observations in part \(k\)
- \(CV_K = \sum_{k=1}^K\frac{n_k}{n}Err_k\)
- \(Err_k = \sum_{i\in C_k}I(y_i\neq\hat{y}_i)/n_k\) (misclassification rate)
- \(n_k\) is the number of observations in group \(k\)
- \(\hat{y}_i\) is the fit for observation \(i\) obtained from the data with the part \(k\) removed
tidymodels
Application Exercise
Create a new
quartofile in your project and addtidymodelsin the name.Load the packages by running the top chunk of R code
tidymodels

- tidymodels is an opinionated collection of R packages designed for modeling and statistical analysis.
- All packages share an underlying philosophy and a common grammar.
Step 1: Specify the model
- Pick the model
- Set the engine
Specify the model
Specify the model
Specify the model
Specify the model
Specify the model
- All available models:
Application Exercise
- Write a pipe that creates a model that uses
lm()to fit a linear regression using tidymodels. Save it aslm_specand look at the object. What does it return?
Hint: you’ll need https://www.tidymodels.org
05:00
Answer
Fit the data
Application Exercise
Answer
parsnip model object
Call:
stats::lm(formula = mpg ~ horsepower, data = data)
Coefficients:
(Intercept) horsepower
39.9359 -0.1578
Call:
lm(formula = mpg ~ horsepower, data = Auto)
Coefficients:
(Intercept) horsepower
39.9359 -0.1578 Get predictions
- Uses the
predict()function - ‼️
new_datahas an underscore - 😄 This automagically creates a data frame
Get predictions
# A tibble: 392 × 10
.pred mpg cylinders displacement horsepower weight acceleration year origin name
* <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct>
1 19.4 18 8 307 130 3504 12 70 1 chevrol…
2 13.9 15 8 350 165 3693 11.5 70 1 buick s…
3 16.3 18 8 318 150 3436 11 70 1 plymout…
4 16.3 16 8 304 150 3433 12 70 1 amc reb…
5 17.8 17 8 302 140 3449 10.5 70 1 ford to…
6 8.68 15 8 429 198 4341 10 70 1 ford ga…
7 5.21 14 8 454 220 4354 9 70 1 chevrol…
8 6.00 14 8 440 215 4312 8.5 70 1 plymout…
9 4.42 14 8 455 225 4425 10 70 1 pontiac…
10 9.95 15 8 390 190 3850 8.5 70 1 amc amb…
# ℹ 382 more rowsWhat does bind_cols do?
Get predictions
# A tibble: 392 × 10
.pred mpg cylinders displacement horsepower weight acceleration year origin name
* <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct>
1 19.4 18 8 307 130 3504 12 70 1 chevrol…
2 13.9 15 8 350 165 3693 11.5 70 1 buick s…
3 16.3 18 8 318 150 3436 11 70 1 plymout…
4 16.3 16 8 304 150 3433 12 70 1 amc reb…
5 17.8 17 8 302 140 3449 10.5 70 1 ford to…
6 8.68 15 8 429 198 4341 10 70 1 ford ga…
7 5.21 14 8 454 220 4354 9 70 1 chevrol…
8 6.00 14 8 440 215 4312 8.5 70 1 plymout…
9 4.42 14 8 455 225 4425 10 70 1 pontiac…
10 9.95 15 8 390 190 3850 8.5 70 1 amc amb…
# ℹ 382 more rowsWhich column has the predicted values?
Application Exercise
03:00
- Edit the code below to add the original data to the predicted data.
Get predictions
# A tibble: 392 × 10
.pred mpg cylinders displacement horsepower weight acceleration year origin name
* <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct>
1 19.4 18 8 307 130 3504 12 70 1 chevrol…
2 13.9 15 8 350 165 3693 11.5 70 1 buick s…
3 16.3 18 8 318 150 3436 11 70 1 plymout…
4 16.3 16 8 304 150 3433 12 70 1 amc reb…
5 17.8 17 8 302 140 3449 10.5 70 1 ford to…
6 8.68 15 8 429 198 4341 10 70 1 ford ga…
7 5.21 14 8 454 220 4354 9 70 1 chevrol…
8 6.00 14 8 440 215 4312 8.5 70 1 plymout…
9 4.42 14 8 455 225 4425 10 70 1 pontiac…
10 9.95 15 8 390 190 3850 8.5 70 1 amc amb…
# ℹ 382 more rowsCalculate the error
- Root mean square error
# A tibble: 1 × 3
.metric .estimator .estimate
<chr> <chr> <dbl>
1 rmse standard 4.89What is this estimate? (training error? testing error?)
Validation set approach
How many observations are in the training set?
Validation set approach
How many observations are in the test set?
Validation set approach
How many observations are there in total?
Validation set approach
- Extract the training and testing data
Validation set approach
# A tibble: 196 × 9
mpg cylinders displacement horsepower weight acceleration year origin name
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct>
1 37.7 4 89 62 2050 17.3 81 3 toyota tercel
2 27 4 97 60 1834 19 71 2 volkswagen mo…
3 22 6 232 112 2835 14.7 82 1 ford granada l
4 16 6 250 100 3781 17 74 1 chevrolet che…
5 25 4 90 71 2223 16.5 75 2 volkswagen da…
6 18 6 232 100 2945 16 73 1 amc hornet
7 38.1 4 89 60 1968 18.8 80 3 toyota coroll…
8 23 4 97 54 2254 23.5 72 2 volkswagen ty…
9 15 8 302 130 4295 14.9 77 1 mercury couga…
10 34 4 108 70 2245 16.9 82 3 toyota corolla
# ℹ 186 more rows Application Exercise
- Copy the code below, fill in the blanks to fit a model on the training data then calculate the test RMSE.
06:00
A faster way!
- You can use
last_fit()and specify the split - This will automatically train the data on the
traindata from the split - Instead of specifying which metric to calculate (with
rmseas before) you can just usecollect_metrics()and it will automatically calculate the metrics on thetestdata from the split
A faster way!
set.seed(100)
Auto_split <- initial_split(Auto, prop = 0.5)
lm_fit <- last_fit(lm_spec,
mpg ~ horsepower,
split = Auto_split)
lm_fit |>
collect_metrics()# A tibble: 2 × 4
.metric .estimator .estimate .config
<chr> <chr> <dbl> <chr>
1 rmse standard 4.96 Preprocessor1_Model1
2 rsq standard 0.613 Preprocessor1_Model1A faster way!
set.seed(100)
Auto_split <- initial_split(Auto, prop = 0.5)
lm_fit <- last_fit(lm_spec,
mpg ~ horsepower,
split = Auto_split)
lm_fit |>
collect_metrics()# A tibble: 2 × 4
.metric .estimator .estimate .config
<chr> <chr> <dbl> <chr>
1 rmse standard 4.96 Preprocessor1_Model1
2 rsq standard 0.613 Preprocessor1_Model1What about cross validation?
What about cross validation?
- Instead of
fitwe will usefit_resamples
What about cross validation?
How do we get the metrics out? With collect_metrics() again!
Application Exercise
05:00
- Edit the code below to get the 5-fold cross validation error rate for the following model:
\(mpg = \beta_0 + \beta_1 horsepower + \beta_2 horsepower^2+ \epsilon\)
- What do you think
rsqis?
Answer
Auto_cv <- vfold_cv(Auto, v = 5)
results <- fit_resamples(lm_spec,
mpg ~ horsepower + I(horsepower^2),
resamples = Auto_cv)
results |>
collect_metrics()# A tibble: 2 × 6
.metric .estimator mean n std_err .config
<chr> <chr> <dbl> <int> <dbl> <chr>
1 rmse standard 4.38 5 0.110 Preprocessor1_Model1
2 rsq standard 0.688 5 0.0177 Preprocessor1_Model1 Application Exercise
Fit 3 models on the data using 5 fold cross validation:
\(mpg = \beta_0 + \beta_1 horsepower + \epsilon\)
\(mpg = \beta_0 + \beta_1 horsepower + \beta_2 horsepower^2+ \epsilon\)
\(mpg = \beta_0 + \beta_1 horsepower + \beta_2 horsepower^2+ \beta_3 horsepower^3 +\epsilon\)
Collect the metrics from each model, saving the results as
results_1,results_2,results_3Which model is “best”?
08:00